Biophysical and structural investigation of the regulation of human GTP cyclohydrolase I by its regulatory protein GFRP.
Ebenhoch, R., Bauer, M., Reinert, D., Kersting, A., Huber, S., Schmid, A., Hinz, I., Feiler, M., Muller, K., Nar, H.(2021) J Struct Biol 213: 107691-107691
- PubMed: 33387654
- DOI: https://doi.org/10.1016/j.jsb.2020.107691
- Primary Citation of Related Structures:
7ALQ - PubMed Abstract:
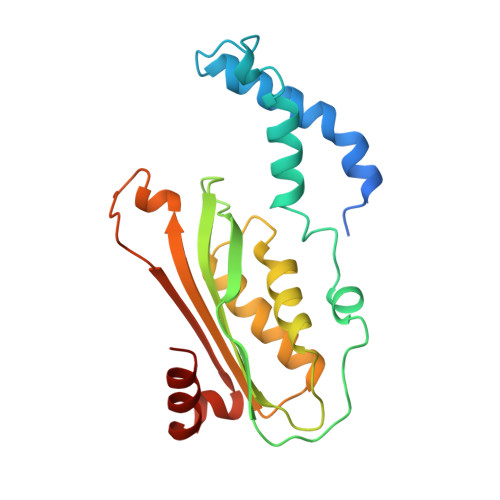
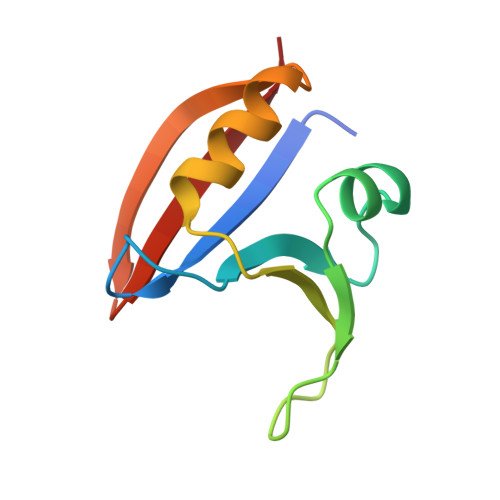
GTP Cyclohydrolase I (GCH1) catalyses the conversion of guanosine triphosphate (GTP) to dihydroneopterin triphosphate (H2NTP), the initiating step in the biosynthesis of tetrahydrobiopterin (BH4). BH4 functions as co-factor in neurotransmitter biosynthesis. BH4 homeostasis is a promising target to treat pain disorders in patients. The function of mammalian GCH1s is regulated by a metabolic sensing mechanism involving a regulator protein, GCH1 feedback regulatory protein (GFRP). Dependent on the relative cellular concentrations of effector ligands, BH4 and phenylalanine, GFRP binds GCH1 to form inhibited or activated complexes, respectively. We determined high-resolution structures of the ligand-free and -bound human GFRP and GCH1-GFRP complexes by X-ray crystallography. Highly similar binding modes of the substrate analogue 7-deaza-GTP to active and inhibited GCH1-GFRP complexes confirm a novel, dissociation rate-controlled mechanism of non-competitive inhibition to be at work. Further, analysis of all structures shows that upon binding of the effector molecules, the conformations of GCH1 or GFRP are altered and form highly complementary surfaces triggering a picomolar interaction of GFRP and GCH1 with extremely slow k off values, while GCH1-GFRP complexes rapidly disintegrate in absence of BH4 or phenylalanine. Finally, comparing behavior of full-length and N-terminally truncated GCH1 we conclude that the disordered GCH1 N-terminus does not have impact on complex formation and enzymatic activity. In summary, this comprehensive and methodologically diverse study helps to provide a better understanding of the regulation of GCH1 by GFRP and could thus stimulate research on GCH1 modulating drugs.
- Boehringer Ingelheim Pharma GmbH & Co. KG, Birkendorfer Str. 65, 88397 Biberach an der Riss, Germany.
Organizational Affiliation: